Generation and analysis of dynamic structural ensembles
Internal dynamics is an intrinsic property of all proteins and is a key determinant of biological function. To analyze states accessible during the internal fluctuations, dynamic structural ensembles, i.e. conformer sets representing the experimentally determined dynamics up to a given time scale can be generated. Such ensembles represent novel models of protein structures and need to be analyzed differently than conventional single-structure models. We constructed the web application CoNSEnsX (Consistency of NMR-derived Structural Ensembles with eXperimental data) allowing fast, simple and convenient assessment of the correspondence of the ensemble as a whole with diverse independent NMR parameters available. It is applied for the evaluation of dynamic conformational ensembles resulting from NMR structure determination. The designed CoNSEnsX approach is freely available as a web service at http://consensx.chem.elte.hu.
The emerging role of internal dynamics in protein fold and function also requires new avenues of structure analysis. We analyzed the dynamically restrained conformational ensemble of ubiquitin generated from residual dipolar coupling data, in terms of protruding and buried atoms as well as interatomic distances, using four proximity-based algorithms, CX, DPX, PRIDE and PRIDE-NMR. We found that ubiquitin, this relatively rigid molecule has a highly diverse dynamic ensemble. We also give a detailed evaluation of PRIDE-NMR on a wide dataset and discuss its usage in the light of the features of available NMR distance restraint sets in public databases.
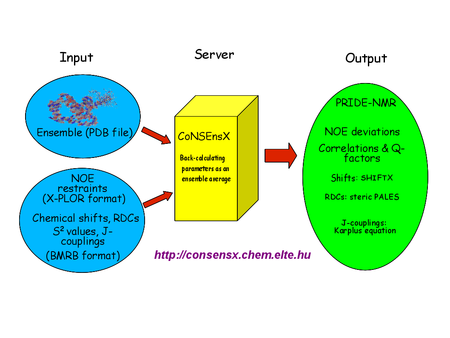
Scheme of the CoNSEnsX approach for the analysis of dynamic structural ensembles
Cooperation with Dr. Sándor Pongor, Protein Structure and Bioinformatics Group, ICGEB Trieste, Italy
Kapcsolódó publikációk
-
András Perczel , Annamária F. Ángyán , Balázs Szappanos , Zoltán Gáspári
CoNSEnsX: an ensemble view of protein structures and NMR-derived experimental data
BMC Structural Biology 10:39. | DOI: 10.1186/1472-6807-10-39. | PMID: 21034466 (2010) Kivonat -
Zoltán Gáspári , Annamária F. Ángyán , Somdutta Dhir , Dino Franklin , András Perczel , Alessandro Pintar , Sándor Pongor
Probing dynamic protein ensembles with atomic proximity measures
Curr. Protein Pept. Sci. 11(7):515-522. | DOI: 10.2174/138920310794109201 | PMID: 20887264 (2010) Kivonat