Structure, dynamics and organization of protein building blocks
An increasing number of diseases, including Alzheimer's, have been found to be a result of the formation of amyloid like aggregates. Consequently, the driving force of the transformation from native to amyloid fold is expected to lie in the protein backbone, which is common to all. By using systematic first principle calculations we show that the ß-pleated sheet structure (building block of amyloid fibers) is thermodynamically the most stable supramolecular arrangement among all of the possible peptide dimers and oligomers, both in vacuum and in aqueous environments. Our study provides a quantum-level explanation of why proteins can take the amyloid state, when local structural preferences are jeopardized and why it is the prefered structure.
We have investigated the stability of fully optimized collagen triple helices and ß-pleated sheets by using ab initio and DFT calculations and have determined their secondary structural preference depending on their amino acid composition. Our method could be used in rational molecular design. Connected to this project, the analysis of the conformation of the Pro-Pro motif was also investigated. The Pro-Pro units are abundant in collagen, in ligand motifs binding to SH3 domains as well as in vital enzymes such as DNA glycosylase and thrombin. The calculated stability data are in good agreement with NMR and X-ray crystallography determined natural abundances.
Since amino acid residues are the simplest building blocks of peptides, the investigation of their conformational properties could help us in understanding their roles in protein folding. We have carried out spectroscopic analysis (IR and VCD) of the simplest chiral and achiral representatives i.e. acetyl-N-methyl-alanine and acetyl-N-methyl-glycine in different solvents as well as in the Ar and Kr matrices. Depending on the conditions different conformers were identified. Our study on the conformational change from extended to 3 10 -helical structure of the simplest oligopeptides (Gly)10 and (Ala)10 revealed the strong side chain-dependence of the internal entropy of folding. It is argued that the difference in information accumulation is related to chirality.
Since the secondary structure elements are known to play a key role in stabilizing the 3D-fold of proteins, for the design of non-natural proteins composed of ß-amino acid residues the construction of suitable secondary structural elements seems to be important. We found the enzyme-resistant non-natural ß-peptides have a remarkably higher affinity to form nanotubes than alpha-peptides. ß-nanotubes can clearly form more diverse secondary structural elements (Fig. 4).
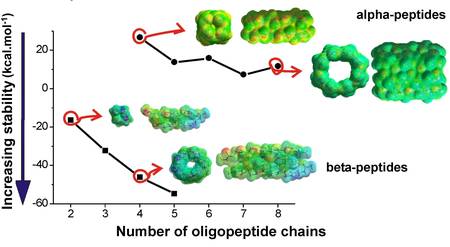
Figure 4. Comparison of the stabilities of peptide nanotubes built from a-amino acid and b-amino acid residues.
For studying the structural basis of tertiary structure folding, Tc5b, a 20-residue long miniprotein was selected. Structural elements influencing conformational stability were studied by NMR methods. We have designed variants with improved folding properties, clearly demonstrating a greater compactness and improved thermal resistance, features typical of globular proteins.