31P-NMR of proteins
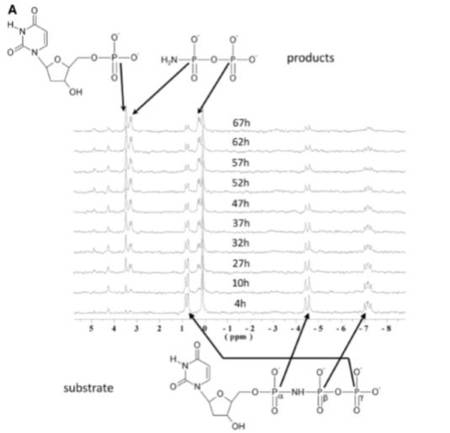
We studied the dUTPase-catalyzed hydrolysis of dUTP, an incorrect DNA building block, to elaborate the mechanistic details at high resolution. Combining mass spectrometry analysis of the dUTPase-catalyzed reaction carried out in and quantum mechanics/molecular mechanics (QM/MM) simulation, we showed that the nucleophilic attack occurs at the α-phosphate site. 31P-NMR spectroscopy analysis confirms the site of attack and shows the capability of dUTPase to cleave the dUTP analogue α,β-imido-dUTP, containing the imido linkage usually regarded to be non-hydrolyzable. We identified a novel enzyme-product complex structure with theis combination of diverse structral methods along with numerous X-ray crystal structures of dUTPase and nucleoside phosphate complexes.
Tubulin polymerization promoting protein/p25 (TPPP/p25) modulates the dynamics and stability of the microtubule system and plays crucial role in the myelination of oligodendrocytes. We showed that the Zn2+-induced change in the TPPP/p25 structure modified its interaction with tubulin and GTP coupled with functional consequences: the TPPP/p25-promoted tubulin polymerization increased, while the TPPP/p25-catalyzed GTPase activity decreased as detected by turbidimetry and by malachite green phosphate release assays and31P-NMR measurements. We also demonstrated by multinuclear NMR that the extended disordered segments of TPPP/p25 are localized at the N- and C-terminals straddling a flexible region.
31P-NMR relaxation measurements were used to obtain detailed information on lipid head-group dynamics, we explored the effect of two topologically different membrane-interacting peptides on bicelles containing either dimyristoylphosphocholine (DMPC), or a mixture of DMPC and dimyristoylphosphoglycerol (DMPG), and dihexanoylphosphocholine (DHPC). KALP21 is a model transmembrane peptide, designed to span a DMPC bilayer and dynorphin B is a membrane surface active neuropeptide.
Myosin basal ATPase cycle for hydrolysis of ATP was also investigated by 31P-NMR to obtain kinetic data.
Cooperation with
Dr. Beáta Vértessy, Department of Applied Biotechnology and Food Sciences, Budapest University of Technology and Economics, Budapest
Dr. Judit Ovádi, Institute of Enzymology, Biological Research Center, Hungarian Academy of Sciences, Budapest
Dr. András Málnási-Csizmadia, Department of Biochemistry, Eötvös Loránd University, Budapest
Dr. Lena Mäler, Department of Biochemistry and Biophysics, Stockholm University, Stockholm, Sweden
Kapcsolódó publikációk
-
Máté Gyimesi , Bálint Kintses , Andrea Bodor , András Perczel , Stefan Fischer , Clive R. Bagshaw , András Málnási-Csizmadia
The mechanism of the reverse recovery step, phosphate release, and actin activation of Dictyostelium myosin II
J. Biol. Chem. 283:8153-8163. (2008) Kivonat -
Zotter A , Oláh J , Hlavanda E , Bodor A , Perczel A , Szigeti K , Fidy J , Ovádi J
Zn(2+)-induced rearrangement of the disordered TPPP/p25 affects its microtubule assembly and GTPase activity.
BIOCHEMISTRY 50:(44) pp. 9568-9578. (2011) Kivonat -
Zotter A , Bodor A , Oláh J , Hlavanda E , Orosz F , Perczel A , Ovádi J
Disordered TPPP/P25 Binds GTP and Displays Mg(2+)-Dependent Gtpase Activity.
FEBS LETTERS 585:(5) pp. 803-808. (2011) Kivonat -
Orsolya Barabás , Vince Grolmusz , Zoltán Szabadka , Imre Zagyva , Zoltán Kele , Edina Rosta , András Perczel , Andrea Bodor , Veronika Németh , Matthias Wilmanns
Catalytic mechanism of α-phosphate attack in dUTPase is revealed by X-ray crystallographic snapshots of distinct intermediates, <sup>31</sup>P-NMR spectroscopy and reaction path modelling
Nucleic Acids Res. DOI: 10.1093/nar/gkt756 | PMID: 23982515 (2013) Kivonat -
Andrea Bodor , Katalin E. Kövér , Lena Mäler
Membrane interactions in small fast-tumbling bicelles as studied by 31P NMR
BBA Biomembranes 1848(3):760-766. | DOI: 10.1016/j.bbamem.2014.12.001 | PMID: 25497765 (2015) Kivonat