Structure, dynamics and activiy of small serine protease inhibitors
SGCI (S. gregaria chymotrypsin inhibitor) and SGTI (S. gregaria trypsin inhibitor) are small, 35-residue peptides isolated from the desert locust, Schistocerca gregaria, with intriguing taxon specificity: SGTI is specific for arthropod proteases while SGCI is an excellent inhibitor on both mammalian and arthropodal enzymes. Both proteins are highly flexible on the ps-ns time scale of motion. However, the internal dynamics of the two closely related molecules are different. We suggested that the dynamic properties of these inhibitors is one of the factors that determine their specificity.
Conformational ensembles reflecting the fast time-scale dynamics of these canonical inhibitors include structures highly similar to the enzyme-bound state. Thus, rather than being classic examples of the rigid lock-and-key mechanism as widely accepted, these inhibitors rather exhibit nanosecond-timescale conformer selection. Thus, fast timescale dynamics enables the association rate to be solely diffusion-controlled just like in the rigid-body model.
Complexation of SGCI with bovine chymotrypsin was also investigated by NMR spectroscopy. Comparison of the free and complexed inhibitor structures revealed that most, but not all, of the observed chemical shift changes can be attributed to minor structural transitions. We suggested that the classical 'scaffold + binding loop' model of canonical inhibitors might not be fully valid for the inhibitor family studied. In our view, this feature allows for the emergence of both taxon-specific and nontaxon-specific inhibitors in this group of small proteins besides the dynamical properties.
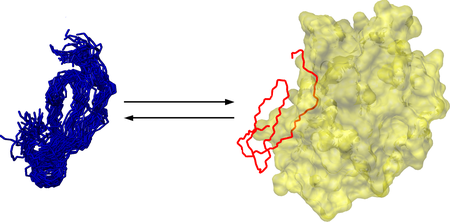
Scheme of nanosecond-timescale conformational selection in SGCI
Cooperation with Prof. Dr. László Gráf, Department of Biochemistry, Eötvös Loránd University, Budapest
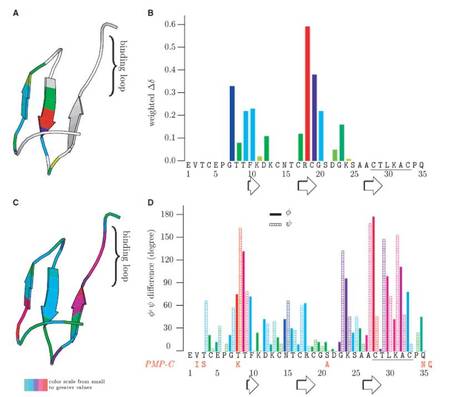
Each family of reversible serine protease inhibitors has a distinct representative structure but contains a surface loop that adopts the same, canonical conformation in the enzyme–inhibitor complex. We tested the interscaffolding additivity theory by randomizing all canonical loop positions of SGCI and SPINK1. We characterized the stability and overall structural and dynamical changes of the inhibitors with DSC, CD, NMR and MD simulations. Our findings disprove interscaffolding additivity and show that loop and scaffold form one integrated unit that needs to be coevolved to provide high-affinity inhibition.
Structures determined
SGCI: [1KGM] SGCI[L30K, K31M]: [1KIO] SGTI: [1KJ0]
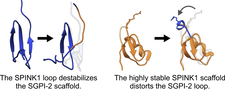
Surface loops of reversible serine protease inhibitors do not act independently of the scaffold
Cooperation with:Prof. Dr. László Gráf, and Dr. Gábor Pál, Department of Biochemistry, Eötvös Loránd University, Budapest, Hungary
Related publications
Related publications
-
Zoltán Gáspári , Péter Várnai , Balázs Szappanos , András Perczel
Reconciling the lock-and-key and dynamic views of canonical serine protease inhibitor action
FEBS Lett. 584(1):203-206. | DOI: 10.1016/j.febslet. | PMID: 19932101 (2010) Kivonat -
Zoltán Gáspári , Borbála Szenthe , András Patthy , William M. Westler , László Gráf , András Perczel
Local binding with globally distributed changes in a small protease inhibitor upon enzyme binding
FEBS J. 273:1831-1842. (2006) Kivonat -
Borbála Szenthe , Zoltán Gáspári , Attila Nagy , András Perczel , László Gráf
Same fold with different mobility: backbone dynamics of small protease inhibitors from the desert locust, Schistocerca gregaria
Biochemistry , 43(12): 3376-3384. (2004) Kivonat -
Péter Rapali , László Radnai , Dániel Süveges , Veronika Harmat , Ferenc Tölgyesi , Weixiao Y. Wahlgren , Gábor Pál
Directed evolution reveals the binding motif preference of the LC8/DYNLL hub protein and predicts large numbers of novel binders in the human proteome
PLoS One 6(4):e18818. | DOI: 10.1371/journal.pone.0018818 | PMID: 21533121 (2011) Kivonat