Complex of crayfish trypsin and SGTI
We solved the crystal structure of crayfish trypsin complexed with SGTI, a taxon selective trypsin inhibitor from desert locust Schistocerca gregaria. The SGTI binding region is extended contacting the P12-P5' residues of the inhibitor. Comparison of the structure of SGTI in the complex and its solution structure (our previous NMR studies) reveals two regions undergoing major conformation change allowing shape adaptation of the inhibitor in the complex. The structure represents the Michaelis complex to atomic resolution (1.2 Angstrom) and ensures deeper insight in serine protease catalysis. Comparison with the acyl-enzyme intermediate of a related enzyme (Katona et al. JBC 277, 21962, 2002) reveals that the strength of the enzyme-substrate hydrogen bonds changes during catalysis.
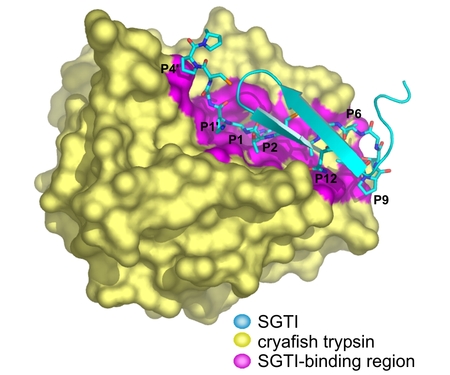
Crayfish trypsin - SGTI complex
Cooperation
Prof. László Gráf,
Department of Biochemistry,
Eötvös Loránd University, Budapest,
Dr. Gergely Katona, Department of Biochemistry, University of Leicester, UK
Structures determined
2F91
Related publications
-
Krisztián Fodor , Veronika Harmat , Csaba Hetényi , József Kardos , József Antal , András Perczel , András Patthy , Gergely Katona , László Gráf
Extended intermolecular interactions in a serine protease canonical inhibitor complex account for strong and highly specific inhibition
J. Mol. Biol. 350:156-169. (2005) Kivonat -
Krisztián Fodor , Veronika Harmat , Richard Neutze , László Szilágyi , László Gráf , Gergely Katona
Enzyme:substrate hydrogen bond shortening during the acylation phase of serine protease catalysis
Biochemistry 45(7):2114-2121. | 10.1021/bi0517133 | PMID: 16475800 (2006) Kivonat