Structure, dynamics and ligand formation of K-Ras oncogenic mutants
K-Ras protein is a membrane-bound small GTPase acting as a molecular switch. It plays a key role in many signal transduction pathways regulating cell proliferation, differentiation, survival, etc.. Its most frequent oncogenic mutants are G12C, G12D, and G12V that have impaired GTPase activity. We reported the resonance assignment of K-Ras wild type, G12C, G12D and G12V proteins’ catalytic G-domain. Simultaneous investigation of these proteins allowed us to perform a comprehensive analysis based on the combined chemical shifts to reveal the effect of mutation at G12 position on structure. The G12C and G12V mutants found to be structurally very similar at the three most important regions of K-Ras (P-loop, Switch-I, Switch-II), while the G12D mutant significantly differs at P-loop and Switch-II from the wildtype as well as G12C and G12V mutants.
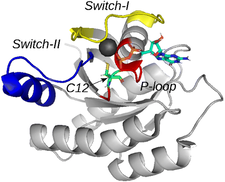
Structure of the wild-type KRas with the three most important regions highlighted
Recently the design of safe and effective covalent drugs has gained significant interest. Remarkable efforts have been made in extending the computational toolkits to include a covalent mechanism of protein targeting. We described a new protocol using tethered and constrained docking in combination with Dynamic Undocking (DUck) for the identification of novel covalent inhibitors. The method was validated on JAK3 and KRas. This fast iterative protocol can be applied to explore a wide chemical space and identify potent targeted covalent inhibitors.
Cooperation with: Dr. György Miklós Keserű, Institute of Organic Chemistry, Hungarian Academy of Sciences, Budapest, Hungary
Related publications
-
Moira Rachman , Andrea Scarpino , Dávid Bajusz , Gyula Pálfy , István Vida , András Perczel , Xavier Barril , György M. Keserű
DUckCov: a Dynamic Undocking-Based Virtual Screening Protocol for Covalent Binders
ChemMedChem 14, 1 – 12 DOI: 10.1002/cmdc.201900078 (2019) Kivonat -
Gyula Pálfy , Dóra K. Menyhárd , András Perczel
Dynamically encoded reactivity of Ras enzymes: opening new frontiers for drug discovery
Cancer and Metastasis Reviews doi.org/10.1007/s10555-020-09917-3 (2024) Kivonat -
Dóra K. Menyhárd , Gyula Pálfy , Zoltán Orgován , István Vida , György M. Keserű , András Perczel
Structural impact of GTP binding on downstream KRAS signaling
Chemical Science DOI: 10.1039/d0sc03441j (2020) Kivonat -
Gyula Pálfy , István Vida , András Perczel
1H, 15N backbone assignment and comparative analysis of the wild type and G12C, G12D, G12V mutants of K‐Ras bound to GDP at physiological pH
Biomolecular NMR Assignments 14, 1-7 | doi.org/10.1007/s12104-019-09909-7 (2020) Kivonat