Novel NMR tools
Degree of denaturation characterized semi quantitatively:
PFG-NMR is used to better understand protein unfolding and to quantitatively characterize the degree of denaturation and aggregation.
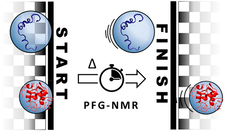
PFG-NMR can distinguish between folded and unfolded states based on their distinct hydrodynamic properties
Real-time pure shift measurements using X-selective BIRD homonuclear decoupling
The applicability of Hα detected spectral approach was probed for proteins to characterize them under physiological conditions.
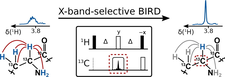
BIRD-based homo-decoupling applied to uniformly isotope-labeled samples
Cooperation with Dr. Burkhard Luy, Institut für Organische Chemie, Karlsruher Institut für Technologie, Karlsruhe, Germany
Semi-quantitative characterization of invisible intermediate stets of proteins
We detected and characterized different intermediate states by T-dependent NMR of the 15N and 13C/15N labeled (mini)proteins, at different pHs and developed a NMR spectra deconvolution technique to characterize the “invisible” intermediate (I-) state(s), beside the fully folded (F-state) and unfolded (U-) state, all in fast exchange. Using nonlinear fitting we derived both the thermodynamic parameters (DH[F–I], Tm[F–I], DCp[F–I] and DH[I–U], Tm[I–U], DCp[I–U]) and the NMR chemical shifts of the conformers of the multistate unfolding process. During the unfolding of Trp-cage miniprotein distinct intermediates evolve: a fast-exchanging intermediate is present under neutral conditions, whereas a slow-exchanging intermediate-pair emerges at acidic pH. Heteronuclear relaxation studies combined with MD simulations revealed the source of backbone mobility and the nature of structural rearrangements during these transitions. The ability to detect structural and dynamic information about folding intermediates in vitro provides an excellent opportunity to gain new insights into the energetic aspects of the energy landscape of protein folding. Our new experimental data offer exceptional testing ground for further computational simulations.

1H,15N HSQC chemical shift changes upon temperature increase of a miniprotein (color of resonances changes gradually from blue (4 C) to purple (54 C).
NMR data based kinetic simulation and thermodynamics
We have completed a systematical analysis of how temperature, pH, presence of charged residues, but most importantly backbone conformation and dynamics affect isomerization rates as determined by NMR in the case of designed Asn/Asp polypeptide-models. We demonstrate that restricted mobility (such as being part of a secondary structural element) may safeguard against Asn to Asp spontaneous isomerization, but this protective factor is most effective in the case of off-pathway folds which can slow the reaction by several magnitudes compared to their on-pathway counterparts. We show that the geometric descriptors of the initial nucleophilic attack of the isomerization can be used to classify local conformation and contribute to the design of stable protein drugs, antibodies or the assessment of the severity of mutations. We concluded that spontaneous deamidation prompted backbone isomerization of Asn/Asp residues resulting in – most cases – the insertion of an extra methylene group into the backbone poses a threat to the structural integrity of proteins.

Time dependent HPLC and NMR information on the backbone Asn/Asp isomerization of a model system
Related publications
-
Petra Rovó , Pál Stráner , András Láng , István Bartha , Kristóf Huszár , László Nyitray , András Perczel
Structural insights into the Trp-cage folding intermediate formation
Chem. Eur. J. 19(8):2628-2640. | DOI: 10.1002/chem.201203764 | PMID: 23319425 (2013) Kivonat -
Erika F. Dudás , Andrea Bodor
Quantitative, Diffusion NMR Based Analytical Tool To Distinguish Folded, Disordered, and Denatured Biomolecules
American Chemical Society 4929-4933 DOI: 10.1021/acs.analchem.8b05617 (2019) Kivonat -
Jens D. Haller , Andrea Bodor , Burkhard Luy
Real-time pure shift measurements for uniformly isotope-labeled molecules using X-selective BIRD homonuclear decoupling
Journal of Magnetic Resonance 64-71 doi.org/10.1016/j.jmr.2019.03.011 (2019) Kivonat -
András Láng , Imre Jákli , Kata Nóra Enyedi , Gábor Mező , Dóra K. Menyhárd , András Perczel
Off-pathway 3D-structure provides protection against spontaneous Asn/Asp isomerization: shielding proteins Achilles heel
Quarterly Reviews of Biophysics 53 | doi:10.1017/S003358351900009X (2020) Kivonat