Design and characterization of Trp-cage miniproteins
Miniproteins are adequate models to study various protein’s 3D-structure modifying parameters, such as temperature, pH, point mutations, H-bonds, salt bridges, molecular packing, etc.. Tc5b, a 20-residue long Trp-cage polypeptide is one of the smallest 3D-folded miniprotein. We reported a systematic investigation of structural factors influencing the stability of Tc5b by solving its solution structure in different environments. Selected variants of Tc5b were designed, prepared and investigated by NMR. Tc6b, (Tc5b_D9E) is a QM designed variant exhibiting an enhanced heat stability: it adopts a stable protein fold at physiological temperature.
Mapping the key residue-residue contacts responsible for Trp-cage fold stability, designing variants and investigating them by NMR and CD spectroscopy showed that the 9-16 salt bridge is integrated into the cooperativity network of the molecule, rather than being an isolated stabilizing factor. Furthermore, we detected and characterized different intermediate states of folding by T-dependent NMR, both at neutral and acidic pHs. We developed an NMR chemical shift deconvolution technique to characterize the invisible, fast exchanging states of the dynamic conformational ensemble of 3D-structures. Using nonlinear fitting methods we obtained both the thermodynamic parameters and the NMR chemical shifts of the pure conformers of such multistate involving folding and unfolding thermodynamic equilibrium. Heteronuclear relaxation studies combined with MD simulations revealed the source of backbone mobility and the nature of structural rearrangements during these T- or pH-induced transitions.
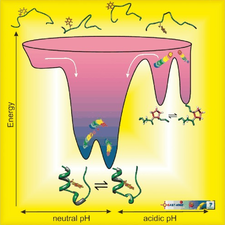
The folding energy landscape of the 20‐residue long Trp‐cage
We designed Exendin-4 (a drug licensed for treating type 2 Diabetes Mellitus) analogues of improved structural characteristics and better water solubility. This rational design started from the 20-amino acid, well-folded Trp-cage miniprotein and involved the step-by-step N-terminal elongation of the Trp-cage head, resulting in finally the 39-amino acid Ex4 analogue, namely E19. 15N relaxation- and diffusion-ordered NMR measurements were completed to investigate their inherent mobility and self-association propensity. Our designed E19 molecule has the same tertiary structure as that of Ex4, but is less prone to oligomerization (aggregation) but preserved its bioactivity. These conditions make E19 a perfect lead compound for further drug discovery.
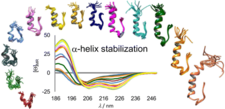
The α-helix-stabilized and step-by-step N-terminal elongated Exendin-4 analogues
We also characterized the amyloid formation of several modified sequence and side-chain phosphorylated Trp-cage variants applying NMR, CD and FTIR spectroscopies and MD simulations. We demonstrated that the native fold is destabilized upon serine phosphorylation, and the resultant highly dynamic structures form amyloid-like ordered aggregates with high intermolecular β-structure content. We proposed a complex aggregation model for these Trp-cage miniproteins.
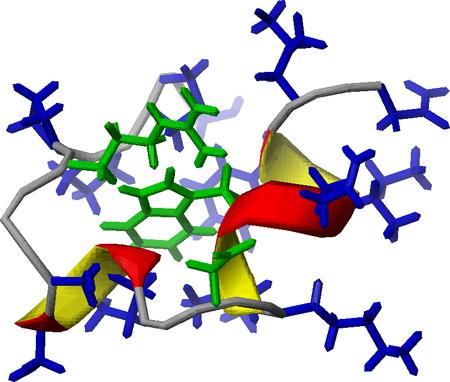
A Trp-cage miniprotein with key stabilizing residues highlighted
Hydration properties of folded (TC5b), semi-folded (TCb5(H+)) and unfolded/disordered (TC5b_N1R) miniproteins were monitored in frozen solutions by wide-line 1H-NMR. Our low temperature denaturation study indicates that freezing of protein solutions proceeds by the gradual selection of the enthalpically most favored states that also minimize the number of bridging waters.
We derived a novel approach to monitor disulfide bond, SS-bond, stabilized Trp-cage protein’s reduction by using combined NMR and ECD spectroscopy. We determined the 3D‐fold characteristics and the associated reduction rate constants of SS-bond reinforced Exenatide mimetics at different experimental conditions. We found that structural, steric and electrostatic factors influence the reduction rate, resulting in magnitude differences in reduction half‐times even for structurally similar, well‐folded model‐protein derivatives.
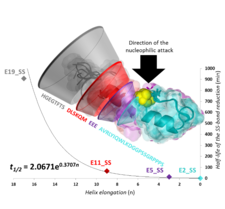
Structural, steric, and electrostatic factors influence the disulfide bond reduction rate.
The amyloid formation mechanism of the folded segment an Exenatide variant was studied by ECD and NMR spectroscopy. Decomposition of the T-, pH-, salt-dependent ECD spectra allowed us to monitor the full range of molecular transformation of amyloidogenesis. Results indicate that the amyloid transition is triggered by the subtle misfolding of the α-helix exposing both aromatic and hydrophobic side-chains to water. These initial misfolded conformational clusters provide the spatial closeness for a transition to the β-structured amyloid nucleus and thus, the amyloid formation process follows a nucleated growth mechanism.
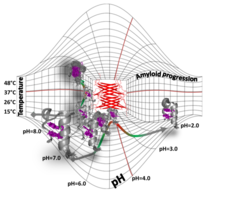
Folded-unfolded-amyloid potential energy surface of Exenatide
- Cooperation with
Dr. Gábor Tóth, Department of Medicinal Chemistry, University of Szeged, Hungary,
- Dr. László Nyitray, Department of Biochemistry, Eötvös Loránd University, Budapest
Related publications
-
József Kardos , Bence Kiss , András Micsonai , Petra Rovó , Dóra K. Menyhárd , János Kovács , Gábor K. Tóth , András Perczel
Phosphorylation as Conformational Switch from the Native to Amyloid State: Trp-Cage as a Protein Aggregation Model
J. Phys. Chem. B 119(7):2946–2955. | DOI: 10.1021/jp5124234 | PMID: 25625571 (2015) Kivonat -
Veronika Harmat , Klarissza Domokos , Dóra K. Menyhárd , Anna Palló , Zoltán Szeltner , Ilona Szamosi , Tamás Beke , Gábor Náray-Szabó , László Polgár
Structure and catalysis of acylamonoacyl peptidase: closed and open subunits of a dimer oligopeptidase
Journal of Biological Chemistry 286:1987-1998. PMID: 21084296 | DOI: 10.1074/jbc.M110.169862 (2011) Kivonat -
Tamás Beke , András Czajlik , Balázs Bálint , András Perczel
A theoretical comparison of self-assembling α- and β-peptide nanostructures: toward design of β-barrel frameworks.
ACS Nano 2:545-553. (2008) Kivonat -
Adriana D. Garro , Ana M. Rodríguez , Javier López Cascales , Botond Penke , Csaba Somlai , András Perczel , Gabriela Feresin , Alejandro Tapia , Beatriz Lima , José A. Bombasaro , Mónica S. Olivella , Ricardo D. Enriz
Penetratin and derivatives acting as antibacterial agents
Chem. Biol. Drug. Des. 82(2):167-177. | DOI: 10.1111/cbdd.12143 | PMID: 23581817 (2013) Kivonat -
Bianka Kótai , György Kardos , Andrea Hamza , Viktor Farkas , Imre Pápai , Tibor Soós
On the Mechanism of Bifunctional Squaramide-Catalyzed Organocatalytic Michael Addition: A Protonated Catalyst as an Oxyanion Hole
Chem. Eur. J. 20(19):5631-5639. | DOI: 10.1002/chem.201304553 | PMID: 24677388 (2014) Kivonat -
Pál Stráner , Nóra Taricska , Mária Szabó , Gábor K. Tóth , András Perczel
Bacterial expression and/or solid phase peptide synthesis of 20-40 amino acid long polypeptides and miniproteins, the case study of Class B GPCR ligands
Curr. Prot. Pept. Sci. 17(2):147-155. DOI: 10.2174/1389203716666151102105215 | PMID: 26521952 (2016) Kivonat -
Nóra Taricska , Dániel Horváth , Dóra K. Menyhárd , Hanna Ákontz-Kiss , Masahiro Noji , Masatomo So , Yuji Goto , Toshimichi Fujiwara , András Perczel
The route from the folded to the amyloid state: exploring the potential energy surface of a drug-like miniprotein
Chemistry A European Journal 26(9), 1968-1978 | doi.org/10.1002/chem.201903826 (2020) Kivonat -
Dániel Horváth , Dóra K. Menyhárd , András Perczel
Protein Aggregation in a Nutshell: The Splendid Molecular Architecture of the Dreaded Amyloid Fibrils
Current Protein & Peptide Science 20(11), 1077-1088 | doi: 10.2174/1389203720666190925102832. (2019) Kivonat -
Petra Rovó , Viktor Farkas , Orsolya Hegyi , Orsolya Szolomájer-Csikós , Gábor K. Tóth , András Perczel
Cooperativity network of Trp-cage miniproteins: probing salt-bridges
J Pept Sci 17(9):610-619. DOI: 10.1002/psc.1377 PMID: 21644245 (2011) Kivonat -
Péter Hudáky , Pál Stráner , Viktor Farkas , Györgyi Váradi , Gábor Tóth , András Perczel
Cooperation between a salt bridge and the hydrophobic core triggers fold stabilization in a Trp-cage miniprotein
Biochemistry 47(3):1007-1016. DOI: 10.1021/bi701371x PMID: 18161949 (2008) Kivonat -
Petra Rovó , Pál Stráner , András Láng , István Bartha , Kristóf Huszár , László Nyitray , András Perczel
Structural insights into the Trp-cage folding intermediate formation
Chem. Eur. J. 19(8):2628-2640. | DOI: 10.1002/chem.201203764 | PMID: 23319425 (2013) Kivonat -
Petra Rovó , Viktor Farkas , Pál Stráner , Mária Szabó , Ágnes Jermendy , Orsolya Hegyi , Gábor K. Tóth , András Perczel
Rational Design of α-Helix-Stabilized Exendin-4 Analogues
Biochemistry 53(22):3540-3552. | DOI: 10.1021/bi500033c | PMID: 24828921 (2014) Kivonat -
Tamás Vajda , András Perczel
Role of water in protein folding, oligomerization, amyloidosis and miniprotein
J. Pept. Sci. 20(10):747-759. | DOI: 10.1002/psc.2671. | PMID: 25098401 (2014) Kivonat -
Dániel Horváth , Nóra Taricska , Ernő Keszei , Pál Stráner , Viktor Farkas , Gábor K. Tóth , András Perczel
Compactness of Protein Folds Alters Disulfide‐Bond Reducibility by Three Orders of Magnitude: A Comprehensive Kinetic Case Study on the Reduction of Differently Sized Tryptophan Cage Model Proteins
ChemBioChem 21(5), 681-695 | doi.org/10.1002/cbic.201900470 (2020) Kivonat